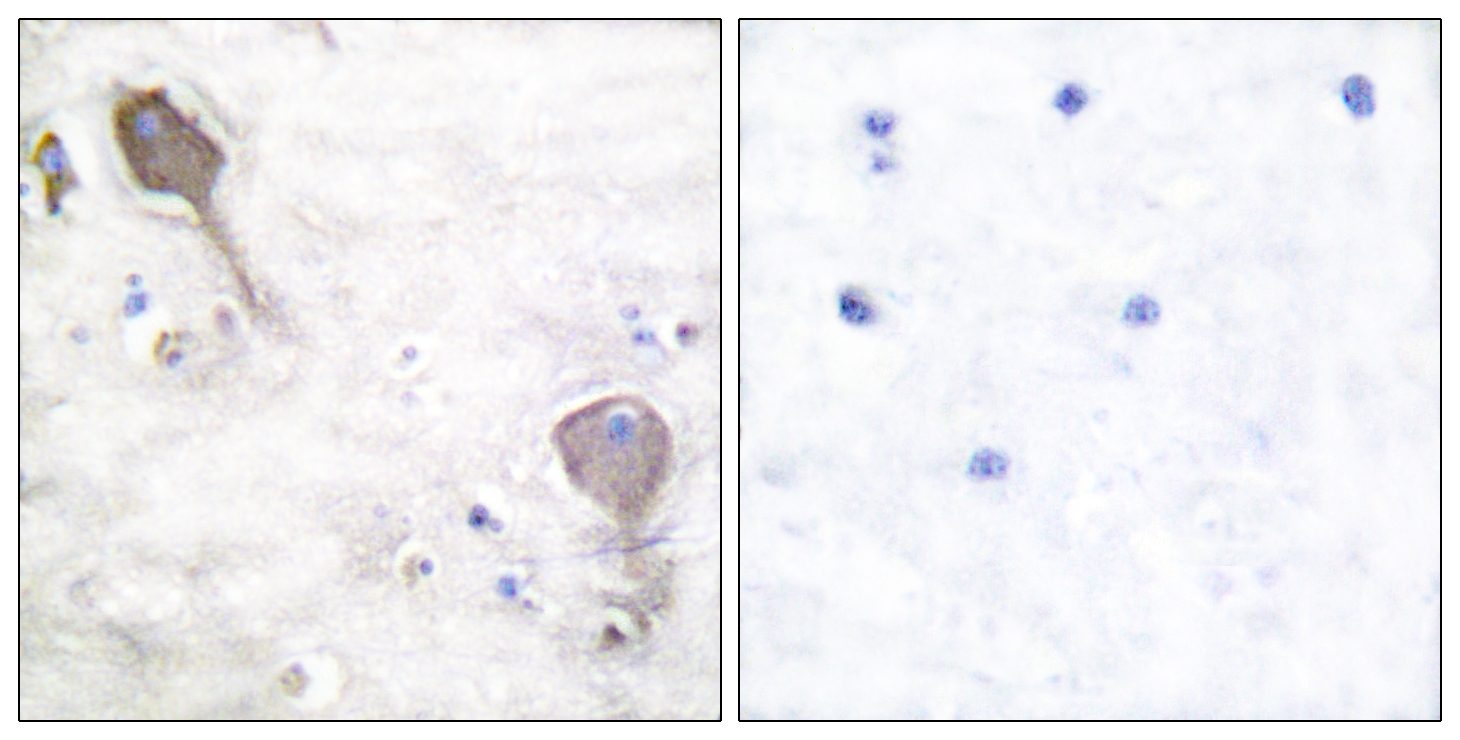
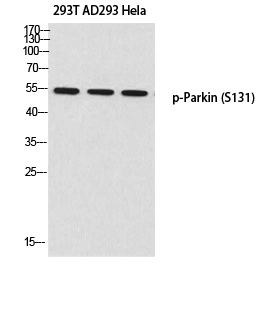
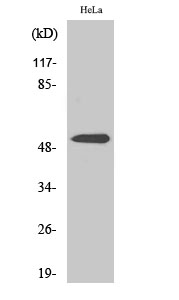
Total PARK2 Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA3353C
- Applications:ELISA
- Reactivity:Human;Mouse;Rat
- Gene Name:
- PARK2
- Human Gene Id:
- 5071
- Human Swiss Prot No:
- O60260
- Mouse Swiss Prot No:
- Q9WVS6
- Rat Swiss Prot No:
- Q9JK66
- Storage Stability:
- 2-8°C/6 months
- Other Name:
- E3 ubiquitin-protein ligase parkin (EC 6.3.2.-) (Parkinson juvenile disease protein 2) (Parkinson disease protein 2)
- Detection Method:
- Colorimetric
- Background:
- disease:Defects in PARK2 are a cause of Parkinson disease (PD) [MIM:168600]. PD is a complex, multifactorial disorder that typically manifests after the age of 50 years, although early-onset cases (before 50 years) are known. PD generally arises as a sporadic condition but is occasionally inherited as a simple mendelian trait. Although sporadic and familial PD are very similar, inherited forms of the disease usually begin at earlier ages and are associated with atypical clinical features. PD is characterized by bradykinesia, resting tremor, muscular rigidity and postural instability, as well as by a clinically significant response to treatment with levodopa. The pathology of PD involves the loss of dopaminergic neurons in the substantia nigra and the presence of Lewy bodies (intraneuronal accumulations of aggregated proteins), in surviving neurons in various areas of the brain.,disease:Defects in PARK2 are the cause of autosomal recessive early onset Parkinson disease 2 (PARK2) [MIM:600116]; also known as early-onset parkinsonism with diurnal fluctuation (EPDF) or autosomal recessive juvenile Parkinson disease (PDJ). PARK2 is symptomatically different in several aspects from idiopathic Parkinson disease, although classic symptoms such as bradykinesia, rigidity and tremor are present. Additional clinical features include early DOPA-induced dyskinesia, diurnal fluctuation of the symptoms, sleep benefit, dystonia and hyper-reflexia. PARK2 is usually characterized by onset before 40, with a mean age at onset of 23.2 years. Pathologically, PARK2 patients show loss of dopaminergic neurons in the substantia nigra, similar to that seen in Parkinson disease; however, Lewy bodies (intraneuronal accumulations of aggregated proteins) are absent.,disease:Defects in PARK2 may be involved in the development and/or progression of ovarian cancer.,domain:The ubiquitin-like domain binds the PSMD4 subunit of 26S proteasomes.,function:Functions within a multiprotein E3 ubiquitin ligase complex, catalyzing the covalent attachment of ubiquitin moieties onto substrate proteins. These substrates include SYT11, CCNE1, GPR37, STUB1, a 22 kDa O-linked glycosylated isoform of SNCAIP and SEPT5. May play a more general role in the ubiquitin proteasomal pathway by participating in the removal and/or detoxification of abnormally folded or damaged protein. Loss of this ubiquitin ligase activity appears to be the mechanism underlying pathogenesis of PARK2. May protect neurons against alpha synuclein toxicity, proteasomal dysfunction, GPR37 accumulation, and kainate-induced excitotoxicity. May play a role in controlling neurotransmitter trafficking at the presynaptic terminal and in calcium-dependent exocytosis. Regulates cyclin E during neuronal apoptosis. May represent a tumor suppressor gene.,miscellaneous:The parkin locus (PRKN), adjacent to the 6q telomere is hyper-recombinable and lies within FRA6E, the third most common fragile site in tumor tissue.,pathway:Protein modification; protein ubiquitination.,PTM:Auto-ubiquitinates in an E2-dependent manner leading to its own degradation.,PTM:S-nitrosylated. The inhibition of PARK2 ubiquitin E3 ligase activity by S-nitrosylation could contribute to the degenerative process in PD by impairing the ubiquitination of PARK2 substrates.,similarity:Belongs to the RBR family. Parkin subfamily.,similarity:Contains 1 IBR-type zinc finger.,similarity:Contains 1 ubiquitin-like domain.,similarity:Contains 2 RING-type zinc fingers.,subcellular location:Co-localizes with SYT11 in neutrites. Co-localizes with SNCAIP in brainstem Lewy bodies.,subunit:Forms an E3 ubiquitin ligase complex with UBE2L3 or UBE2L6. Part of a SCF-like complex, consisting of PARK2, CUL1 and FBXW7. Interacts with SNCAIP. Binds to the C2A and C2B domains of SYT11. Interacts and regulates the turnover of SEPT5. Part of a complex, including STUB1, HSP70 and GPR37. The amount of STUB1 in the complex increases during ER stress. STUB1 promotes the dissociation of HSP70 from PARK2 and GPR37, thus facilitating PARK2-mediated GPR37 ubiquitination. HSP70 transiently associates with unfolded GPR37 and inhibits the E3 activity of PARK2, whereas, STUB1 enhances the E3 activity of PARK2 through promotion of dissociation of HSP70 from PARK2-GPR37 complexes. Interacts with PSMD4 and PACRG. Interacts with LRRK2. Interacts with RANBP2. Interacts with SUMO1 but not SUMO2, which promotes nuclear localization and autoubiquitination.,tissue specificity:Highly expressed in the brain including the substantia nigra. Expressed in heart, testis and skeletal muscle. Expression is down-regulated or absent in tumor biopsies, and absent in the brain of PARK2 patients. Overexpression protects dopamine neurons from kainate-mediated apoptosis.,
- Function:
- neurotransmitter uptake, regulation of neurotransmitter levels, synaptic transmission, dopaminergic, startle response,proteolysis, ubiquitin-dependent protein catabolic process, ubiquitin cycle, cellular amino acid derivative metabolic process, biogenic amine metabolic process, catecholamine metabolic process, neurotransmitter transport, cell-cell signaling, synaptic transmission, nerve-nerve synaptic transmission, behavior, learning or memory, learning,locomotory behavior, adult locomotory behavior, macromolecule catabolic process, catechol metabolic process, amine transport, monoamine transport, organic alcohol transport, dopamine transport, protein ubiquitination, phenol metabolic process, transmission of nerve impulse, modification-dependent protein catabolic process, protein catabolic process, adult behavior, regulation of neurological system process, protein modification by small p
- Subcellular Location:
- Cytoplasm, cytosol . Nucleus . Endoplasmic reticulum . Mitochondrion . Mitochondrion outer membrane . Cell projection, neuron projection . Cell junction, synapse, postsynaptic density . Cell junction, synapse, presynapse . Mainly localizes in the cytosol (PubMed:19029340, PubMed:19229105). Co-localizes with SYT11 in neutrites (PubMed:12925569). Co-localizes with SNCAIP in brainstem Lewy bodies (PubMed:10319893, PubMed:11431533). Translocates to dysfunctional mitochondria that have lost the mitochondrial membrane potential; recruitment to mitochondria is PINK1-dependent (PubMed:24898855, PubMed:18957282, PubMed:19966284, PubMed:23620051). Mitochondrial localization also gradually increases with cellular growth (PubMed:22082830). .
- Expression:
- Highly expressed in the brain including the substantia nigra (PubMed:9560156, PubMed:19501131). Expressed in heart, testis and skeletal muscle (PubMed:9560156). Expression is down-regulated or absent in tumor biopsies, and absent in the brain of PARK2 patients (PubMed:14614460, PubMed:12719539). Overexpression protects dopamine neurons from kainate-mediated apoptosis (PubMed:12628165). Found in serum (at protein level) (PubMed:19501131).
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs