Rad2/FEN1 (Phospho Ser187) rabbit pAb
- Catalog No.:YP1733
- Applications:WB
- Reactivity:Human;Mouse;Rat
- Target:
- FEN-1
- Fields:
- >>DNA replication;>>Base excision repair;>>Non-homologous end-joining
- Gene Name:
- FEN1 RAD2
- Protein Name:
- Rad2/FEN1 (Phospho-Ser187)
- Human Gene Id:
- 2237
- Human Swiss Prot No:
- P39748
- Mouse Swiss Prot No:
- P39749
- Rat Gene Id:
- 84490
- Rat Swiss Prot No:
- Q5XIP6
- Immunogen:
- Synthesized peptide derived from human Rad2/FEN1 (Phospho-Ser187)
- Specificity:
- This antibody detects endogenous levels of Rad2/FEN1 (Phospho-Ser187) at Human, Mouse,Rat
- Formulation:
- Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide.
- Source:
- Polyclonal, Rabbit,IgG
- Dilution:
- WB 1:500-2000
- Purification:
- The antibody was affinity-purified from rabbit serum by affinity-chromatography using specific immunogen.
- Concentration:
- 1 mg/ml
- Storage Stability:
- -15°C to -25°C/1 year(Do not lower than -25°C)
- Other Name:
- Flap endonuclease 1 (FEN-1) (EC 3.1.-.-) (DNase IV) (Flap structure-specific endonuclease 1) (Maturation factor 1) (MF1) (hFEN-1)
- Observed Band(KD):
- 42kD
- Background:
- The protein encoded by this gene removes 5' overhanging flaps in DNA repair and processes the 5' ends of Okazaki fragments in lagging strand DNA synthesis. Direct physical interaction between this protein and AP endonuclease 1 during long-patch base excision repair provides coordinated loading of the proteins onto the substrate, thus passing the substrate from one enzyme to another. The protein is a member of the XPG/RAD2 endonuclease family and is one of ten proteins essential for cell-free DNA replication. DNA secondary structure can inhibit flap processing at certain trinucleotide repeats in a length-dependent manner by concealing the 5' end of the flap that is necessary for both binding and cleavage by the protein encoded by this gene. Therefore, secondary structure can deter the protective function of this protein, leading to site-specific trinucleotide expansions
- Function:
- cofactor:Binds 2 magnesium ions per subunit. They probably participate in the reaction catalyzed by the enzyme. May bind an additional third magnesium ion after substrate binding.,function:Endonuclease that cleaves the 5'-overhanging flap structure that is generated by displacement synthesis when DNA polymerase encounters the 5'-end of a downstream Okazaki fragment. Also possesses 5' to 3' exonuclease activity on niked or gapped double-stranded DNA, and exhibits RNase H activity.,PTM:Acetylated by EP300. Acetylation inhibits both endonuclease and exonuclease activity. Acetylation also reduces DNA-binding activity but does not affect interaction with PCNA or EP300.,similarity:Belongs to the XPG/RAD2 endonuclease family. FEN1 subfamily.,subunit:Interacts with PCNA. The C-terminal domain binds EP300. Can bind simultaneously to both PCNA and EP300.,
- Subcellular Location:
- [Isoform 1]: Nucleus, nucleolus. Nucleus, nucleoplasm. Resides mostly in the nucleoli and relocalizes to the nucleoplasm upon DNA damage.; [Isoform FENMIT]: Mitochondrion .
- Expression:
- Breast,Leukemic T-cell,Lung,
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs
- Products Images
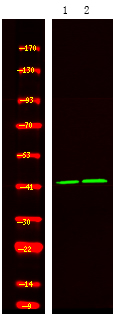
- Western Blot analysis of 1HeLa cell, 2 LPS 100ng/mL 30min treated ,using primary antibody at 1:1000 dilution. Secondary antibody(catalog#:RS23920) was diluted at 1:10000