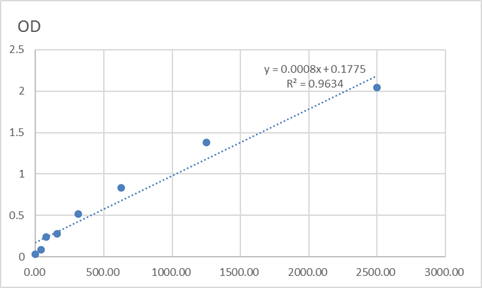
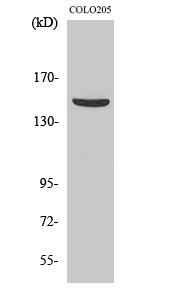
Total PARD3A Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA3829C
- Applications:ELISA
- Reactivity:Human;Mouse;Rat
- Gene Name:
- PARD3
- Human Gene Id:
- 56288
- Human Swiss Prot No:
- Q8TEW0
- Mouse Swiss Prot No:
- Q99NH2
- Rat Swiss Prot No:
- Q9Z340
- Storage Stability:
- 2-8°C/6 months
- Other Name:
- Partitioning defective 3 homolog (PAR-3) (PARD-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2-5) (PAR3-alpha)
- Detection Method:
- Colorimetric
- Background:
- alternative products:Additional isoforms seem to exist. As a matter of fact, alternatively spliced products seem to fall into two broad groups: one group, which includes the longest continuous ORF but which may also include molecules lacking some middle domains, has a single TM element and is likely to be associated with the plasma membrane. The other group lacks a TM domain and thus its members may be secreted,disease:Defects in PKHD1 are the cause of polycystic kidney disease autosomal recessive (ARPKD) [MIM:263200]. ARPKD is a severe form of polycystic kidney disease affecting the kidneys and the hepatic biliary tract. The clinical spectrum is widely variable, with most cases presenting during infancy. The fetal phenotypic features classically include enlarged and echogenic kidneys, as well as oligohydramnios secondary to a poor urine output. Up to 50% of the affected neonates die shortly after birth, as a result of severe pulmonary hypoplasia and secondary respiratory insufficiency. In the subset that survives the perinatal period, morbidity and mortality are mainly related to severe systemic hypertension, renal insufficiency, and portal hypertension due to portal-tract fibrosis.,domain:Contains a conserved N-terminal oligomerization domain (NTD) that is involved in oligomerization and is essential for proper subapical membrane localization.,function:Adapter protein involved in asymmetrical cell division and cell polarization processes. Seems to play a central role in the formation of epithelial tight junctions. Association with PARD6B may prevent the interaction of PARD3 with F11R/JAM1, thereby preventing tight junction assembly. The PARD6-PARD3 complex links GTP-bound Rho small GTPases to atypical protein kinase C proteins.,function:May be a receptor protein that acts in collecting-duct and biliary differentiation.,miscellaneous:Antibodies against PARD3 are present in sera from patients with cutaneous T-cell lymphomas.,PTM:Phosphorylated by PRKCZ. EGF-induced Tyr-1127 phosphorylation mediates dissociation from LIMK2.,sequence caution:Contaminating sequence. Potential poly-A sequence.,similarity:Belongs to the PAR3 family.,similarity:Contains 12 IPT/TIG domains.,similarity:Contains 3 PDZ (DHR) domains.,similarity:Contains 9 PbH1 repeats.,subcellular location:Localized along the cell-cell contact region. Colocalizes with PARD6A and PRKCI at epithelial tight junctions. Colocalizes with the cortical actin that overlays the meiotic spindle during metaphase I and metaphase II.,subunit:Interacts with PARD6A and PARD6B. Isoform 2, but not at least isoform 3 interacts with PRKCZ. Interacts with PRCKI (By similarity). Part of a complex with PARD6A or PARD6B, PRKCI or PRKCZ and CDC42 or RAC1. Interacts with F11R/JAM1 (By similarity). Component of a complex whose core is composed of ARHGAP17, AMOT, MPP5/PALS1, INADL/PATJ and PARD3/PAR3. Interacts with LIMK2.,tissue specificity:Predominantly expressed in fetal and adult kidney. Also present in the adult pancreas, but at much lower levels. Detectable in fetal and adult liver. Rather indistinct signal in fetal brain.,tissue specificity:Widely expressed.,
- Function:
- cell morphogenesis, cell morphogenesis involved in differentiation, protein complex assembly, cell cycle, cell adhesion,establishment or maintenance of cell polarity, cell surface receptor linked signal transduction, G-protein coupled receptor protein signaling pathway, activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway, axonogenesis, asymmetric cell division, cell-cell adhesion, regulation of phosphate metabolic process, biological adhesion, cell projection organization, neuron differentiation, neuron projection development,activation of protein kinase activity, cellular component morphogenesis, cell part morphogenesis, positive regulation of kinase activity, regulation of phosphorylation, homeostatic process, positive regulation of catalytic activity, regulation of kinase activity, macromolecular complex subunit organization, positive regula
- Subcellular Location:
- Cytoplasm . Endomembrane system . Cell junction . Cell junction, tight junction . Cell junction, adherens junction . Cell membrane . Cytoplasm, cell cortex . Cytoplasm, cytoskeleton . Localized along the cell-cell contact region. Colocalizes with PARD6A and PRKCI at epithelial tight junctions. Colocalizes with the cortical actin that overlays the meiotic spindle during metaphase I and metaphase II. Colocalized with SIRT2 in internode region of myelin sheath (By similarity). Presence of KRIT1, CDH5 and RAP1B is required for its localization to the cell junction. .
- Expression:
- Widely expressed.
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs